To effectively develop countermeasures for emerging and re-emerging (viral) pandemics, a significant collaborative effort between industry, government and academia, at a global level, is necessary. Scientific countermeasures include the development of diagnostic tests, a strong research effort in vaccine and therapeutic development and the constant surveillance of virus strains for structural variations. For example, the WHO’s (1) Global Influenza Program (GIP) (2) collects global epidemiological and virology data on circulating influenza virus strains; on careful watch for any signs of emerging virulent strains (3). Data from here, sets the global standard for selection of strains for flu vaccine production (4), twice per year, for immunity against these selected strains. This is vital as the influenza virus genome (or any virus genome) can change via antigenic drifts and shifts (5,6). The result is a change or mutation(s) in surface proteins (antigens); antigens are molecules recognized by the immune system and capable of triggering an immune response (including antibody production) (7).
Unprecedented is the global scientific impact as the race for a vaccine and therapeutic development against the novel coronavirus (SARS-CoV-2) proceeds. This includes, testing of old candidates (8), developing new ones and improved diagnostics. To put this transformation into perspective it is necessary to revisit the past. The 1918 influenza pandemic was the deadliest in the history of the world (9). Initial vaccines were made from bacterial formulations as some experts believed that bacteria caused the infection (10). It was deduced until the 1930s that the causative agent was a virus; the first effective experimental influenza vaccines were then tested (11). To date, there are no approved vaccines or therapeutics against SAR-CoV-2 (disease: COVID-19) however there are 67 candidates in pre-clinical and three in clinical evaluations across the globe (12) . Candidate therapeutics include antibodies and, repurposed and in-development antiviral drugs (13). Over 200 clinical trials are ongoing not only with candidate vaccines and therapeutics (13), but also with traditional medicines and plasma from recovered COVID-19 patients (14); the idea is that plasma comprises antibodies which would be able to mitigate infection by blocking virus attachment to target cells, neutralizing the virus (15).
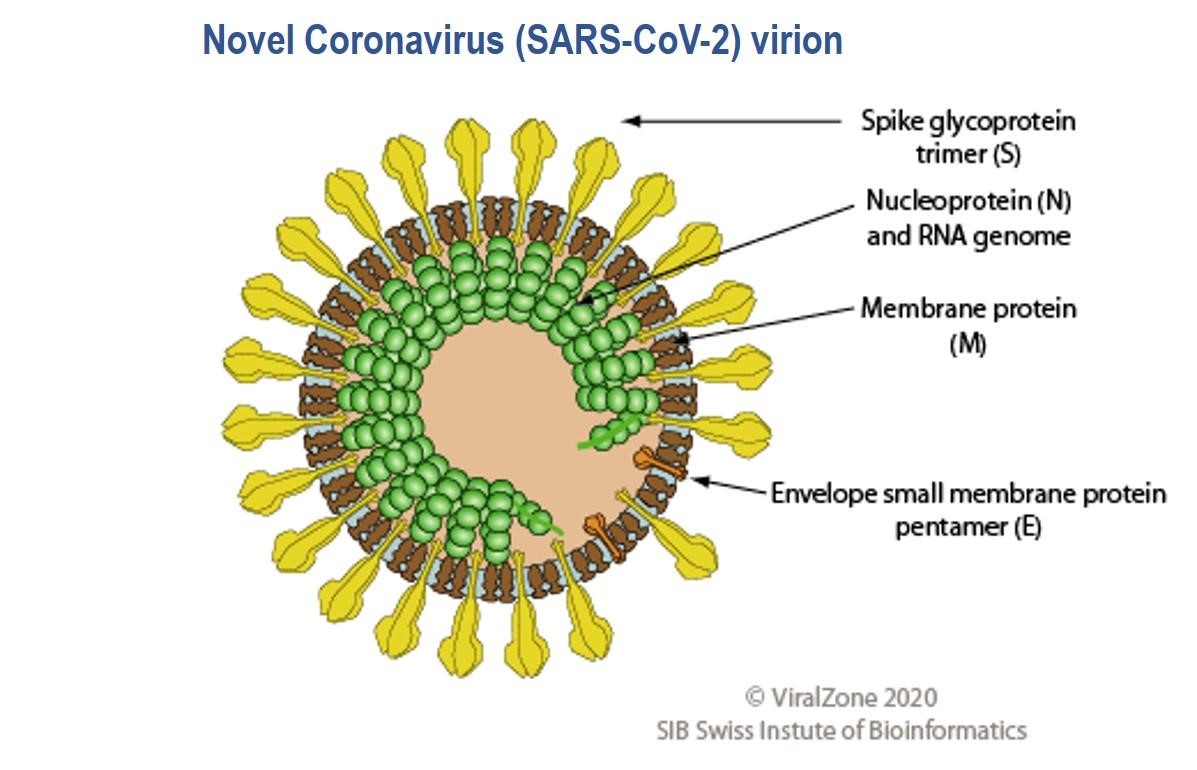
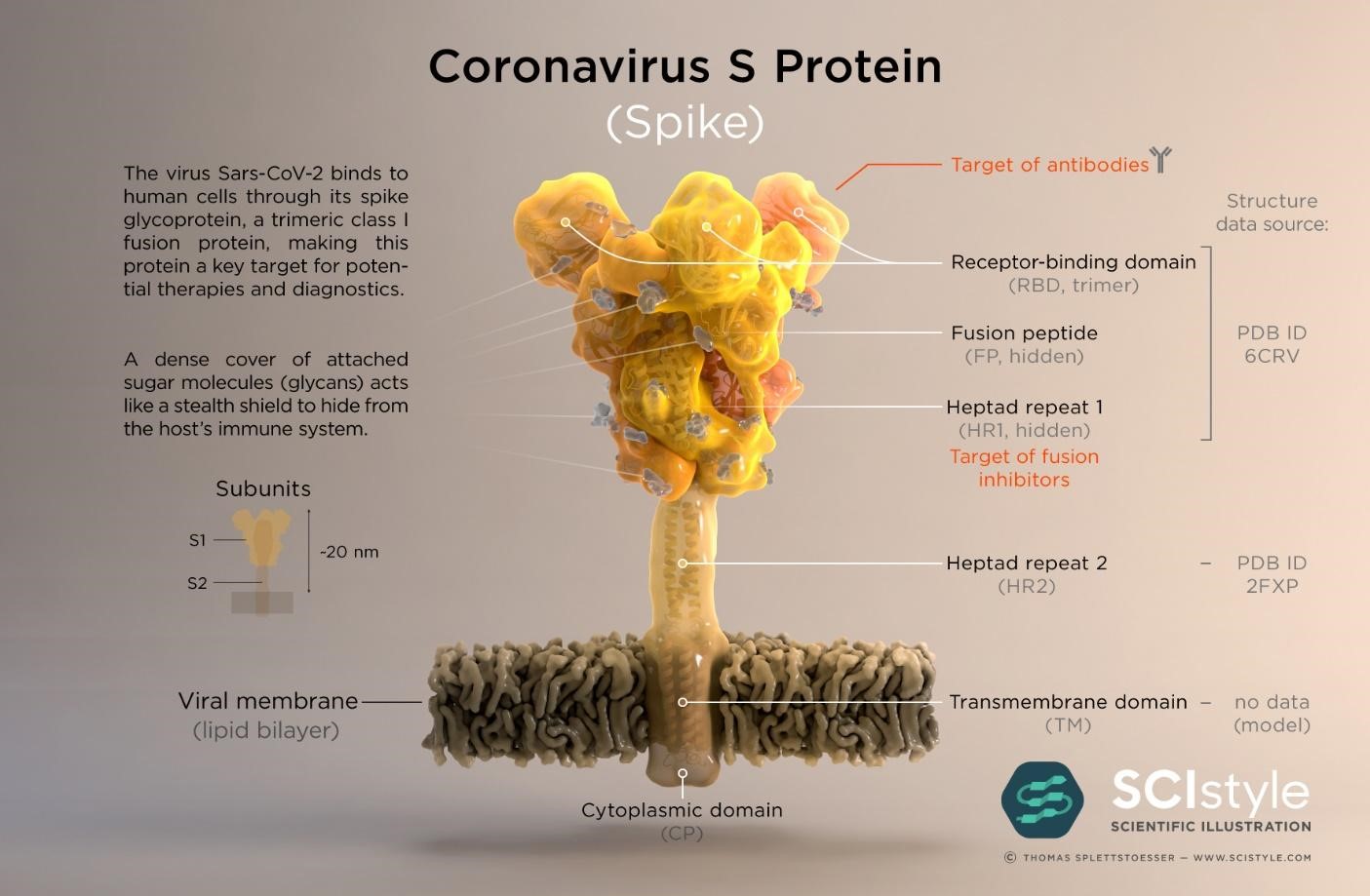
The SARS-CoV-2 virus genome was sequenced and made available on January 10th 2020 less than a month after the WHO was informed of pneumonia cases from an unknown disease in Wuhan (16,17); the causative agent was identified as the SARS-CoV-2 virus on January 7th. Genes which encode twenty nine proteins have been identified including the CoV spike (S) glycoprotein surface antigen (18,19); research shows it is a target for antibodies and therefore a target for diagnostics, vaccines and therapeutic development (20). For the 1918 influenza pandemic, scientists were only able to sequence part of the influenza virus genome 79 years later (21); it was done using preserved lung tissue from a victim who died in 1918 pandemic. In 1999, sequencing of the full length haemagglutinin (HA) gene (encodes HA, a surface protein) was achieved, pioneering vaccine and drug development with HA as a target (22,23). By 2005, with advancement in genomics technology, the entire genome of the 1918 virus was sequenced (24). This allowed the live virus to be reconstructed at the CDC and fully studied to determine properties that contributed to pathogenicity and virulence, further aiding vaccine and drug development (25,26). To date, at the CDC, SARS-CoV-2 has been grown in cell culture for similar research purposes (27)
Real Time reverse transcription-Polymerase Chain Reaction (rRT-PCR) is the main technique used to identify SARS-CoV-2 (28); what most media and public reports on testing is based upon. Half a Nobel Prize was awarded in (1993) for the development of the PCR method (29). SARS-CoV-2 is detected by its specific viral (genome) signature in nasal secretion samples usually taken from the back of the nose or throat. Biopharma researchers are working on different versions of this test and different instrumentation (30). A main limitation of this test is that it only detects SAR-CoV-2 in samples taken from patients with active infections and not from those who have recovered. Hence the need for serological tests (already in development) which would allow a more comprehensive tracking of COVID-19 progression beyond the infectious stage (31). Serological tests measure the amount of antibodies present in the blood as the body responds to an infection (the immune response) and as the disease progresses. It would be an invaluable tool when used in combination with the diagnostic rRT-PCR test in assessing how widespread COVID-19 is and who may have developed immunity and for how long.
Overall, research and developments efforts targeted towards COVID-19 have moved at a significant pace, supported by technological advancement in genomics, proteomics and analytical methodologies. Although a lot has been achieved in a short period of time, I think the challenge for scientists is to provide high quality results at a rapid pace while trying to understand essential fundamental science; the challenge to meet societal needs and expectations and endure scrutiny without compromising scientific standards.
References
1) https://www.who.int/blueprint/priority-diseases/key-action/Novel-Coronavirus_Landscape_nCoV-4april2020.pdf?ua=1
2) https://www.who.int/influenza/surveillance_monitoring/en/
3) https://www.jci.org/articles/view/44947
4) https://www.cdc.gov/flu/prevent/keyfacts.htm
5) https://www.cdc.gov/flu/about/viruses/types.htm
6) https://www.cdc.gov/flu/about/viruses/change.htm
7) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2923430/
8) https://www.statnews.com/2020/03/18/who-to-launch-multinational-trial-to-jumpstart-search-for-coronavirus-drugs/
9) https://www.who.int/influenza/pandemic-influenza-an-evolving-challenge/en/
10) https://www.aai.org/AAISite/media/About/History/Articles/AAI_History_007-(1).pdf
11) https://www.pnas.org/content/pnas/111/34/12283.full.pdf
12) https://www.who.int/blueprint/priority-diseases/key-action/Novel_Coronavirus_Landscape_nCoV_11April2020.PDF?ua=1
13) https://www.who.int/blueprint/priority-diseases/key-action/Coronavirus_Roadmap_V9.pdf?ua=1
14) https://clinicaltrials.gov/ct2/show/NCT04323800?term=convalescent+plasma&cond=COVID-19&draw=2#contacts
15) https://www.hindawi.com/journals/ab/2014/157895/
16) https://www.ncbi.nlm.nih.gov/nuccore/MN908947
17) https://www.who.int/blueprint/priority-diseases/key-action/novel-coronavirus/en/
18) https://cen.acs.org/biological-chemistry/infectious-disease/know-novel-coronaviruss-29-proteins/98/web/2020/04
19) https://science.sciencemag.org/content/367/6483/1260
20) https://www.sciencedirect.com/science/article/pii/S0092867420302622
21) https://science.sciencemag.org/content/275/5307/1793/tab-pdf
22) https://www.pnas.org/content/96/4/1651/tab-article-info
23) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6563292/
24) https://www.sciencedirect.com/science/article/pii/S0092867413011410
25) https://science.sciencemag.org/content/310/5745/77.full
26) https://www.cdc.gov/flu/about/qa/1918flupandemic.htm
27) https://www.cdc.gov/coronavirus/2019-ncov/php/grows-virus-cell-culture.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fabout%2Fgrows-virus-cell-culture.html
28) https://www.cdc.gov/coronavirus/2019-ncov/lab/index.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Flab%2Frt-pcr-detection-instructions.html
29) https://www.nobelprize.org/prizes/chemistry/1993/summary/
30) https://abbott.mediaroom.com/2020-03-27-Abbott-Launches-Molecular-Point-of-Care-Test-to-Detect-Novel-Coronavirus-in-as-Little-as-Five-Minutes
31) https://www.cdc.gov/coronavirus/2019-ncov/php/testing.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fabout%2Ftesting.html